BiocSwirl is a series of in-depth swirlify generated courses used to teach bioinformatics workflows in R/Bioconductor using an interactive and easy to digest format.
About
There have been many resources available to interdisciplinary researchers for years now in the form of Rbookdown tutorials, in person workshops, and Youtube videos. However, each method of learning has its pros and cons - for blogpost and video type tutorials we have the inability to troubleshoot and raise our hands, for in-person workshops we have time limitations. Bioinformatics has grown in adoption in many traditional non-computer science fields that lack the time and energy to go truly in depth past copying and pasting code found online. Swirlstats and terminal-based Python courses such as browser-based Codeacademy, have shown to be efficient course structures to teach data science and statistics to complete beginners.
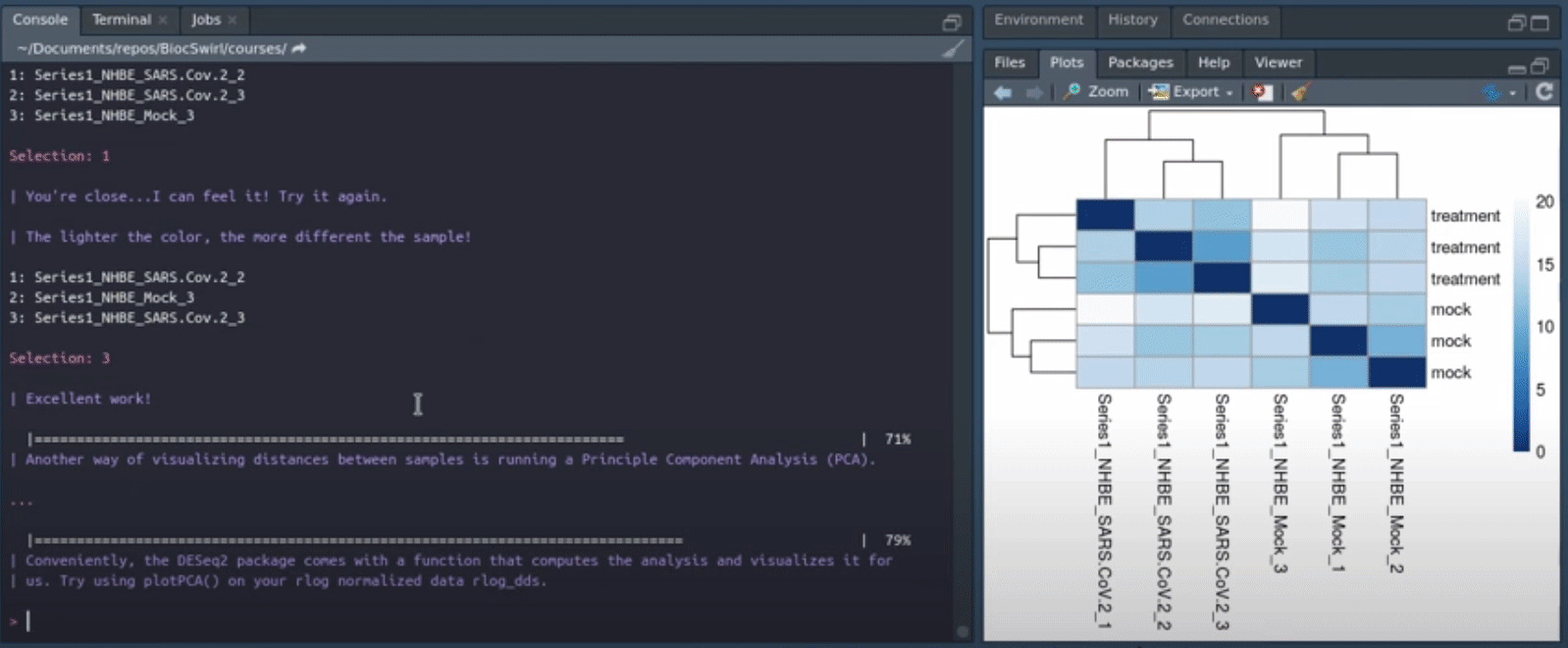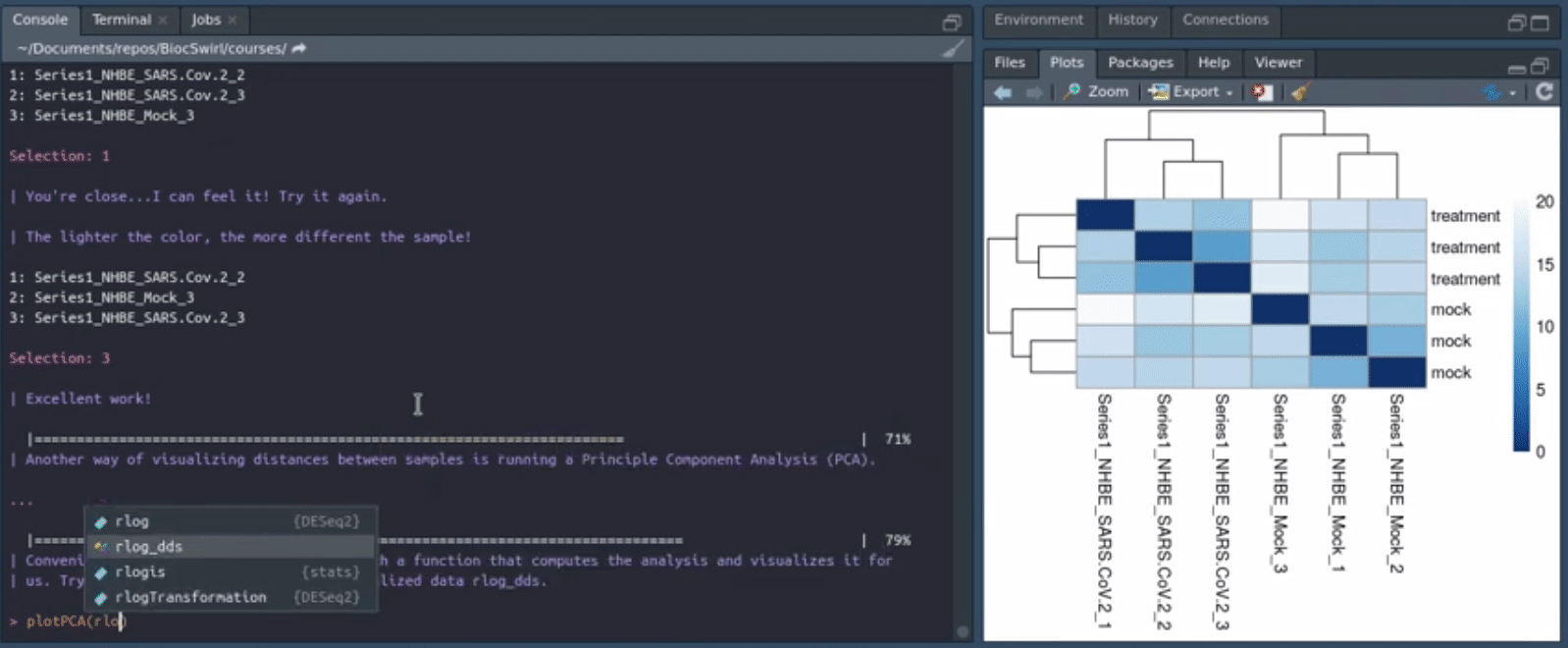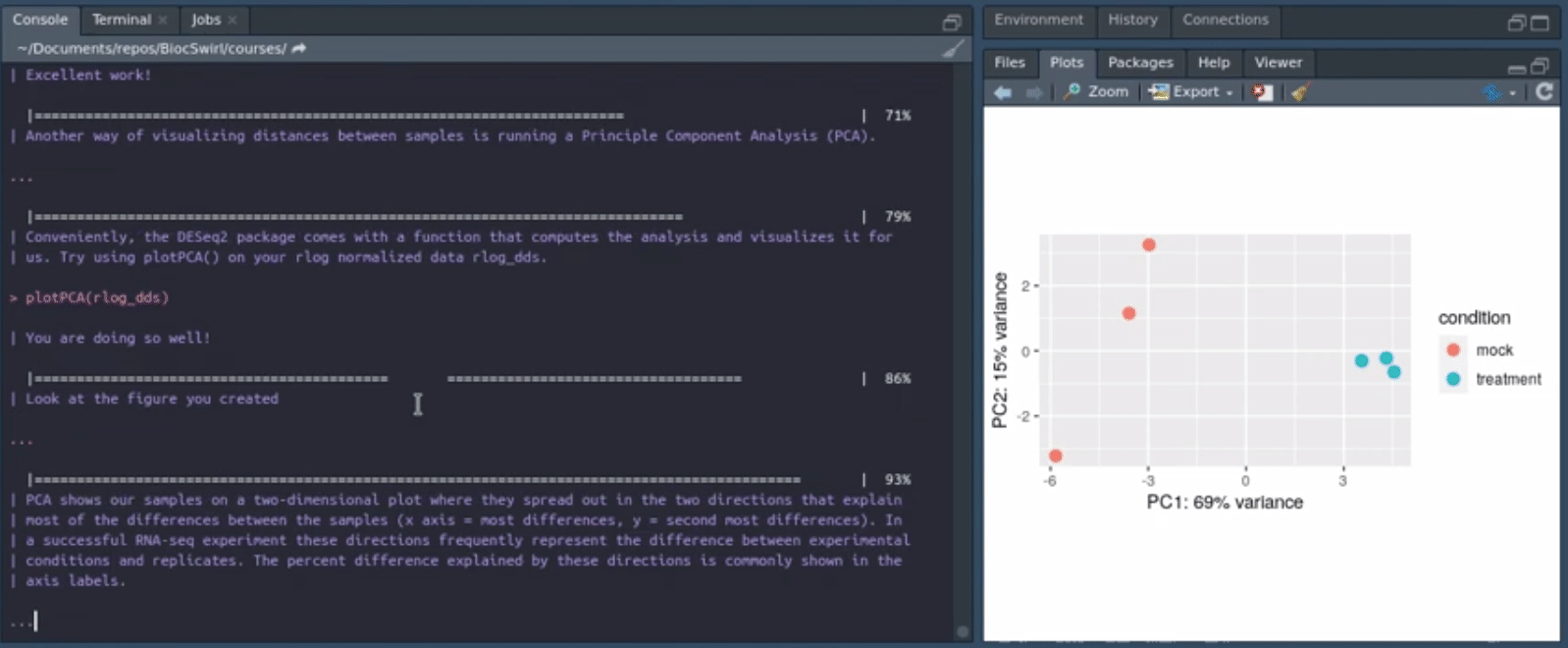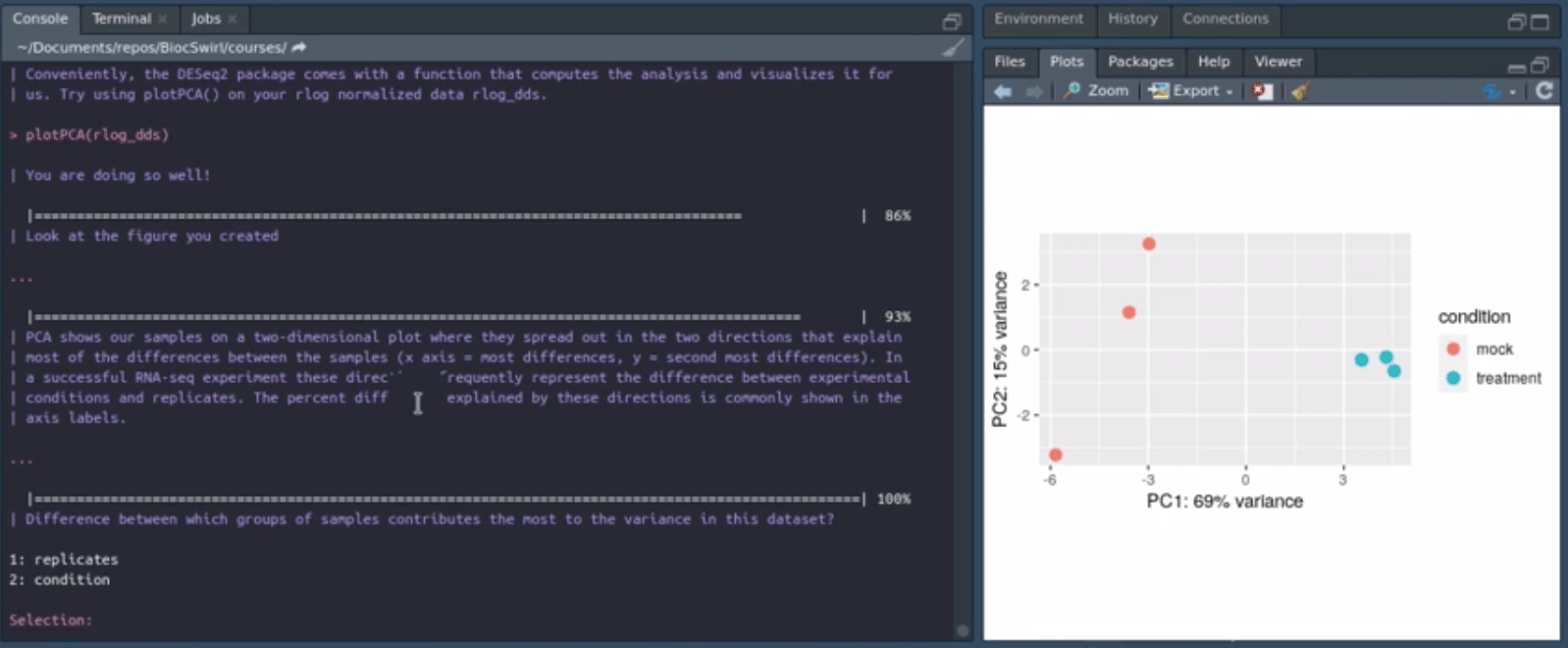
We develop swirlify()-style console/terminal based courses for the purposes of teaching bioinformatics workflow steps and good coding practices. The philosophy is to keep the GUI use and application switching as minimal as possible but the content itself very rich and informative. See also BiocTerm (still in development) for our interface companion to the course materials. The interface (BiocTerm) and R package (BiocSwirl) can be used independently of each other but are best used together for people who are interested in making the most of their learning experience.
Installation
Use the following code to install BiocSwirl via R:
# install.packages("devtools")
# library(devtools)
devtools::install_github("biocswirl-dev-team/BiocSwirl")
library(BiocSwirl)
Alternatively, the BiocSwirl courses can be installed independently. Please follow these instructions to manually install our courses: https://github.com/swirldev/swirl_courses/wiki/Legacy-Manual-Install-Instructions-for-Swirl-Courses
Submission to Bioconductor pending.
Course Offerings
We're currently offering five different interactive courses: Introduction to Data Science for Life Scientists, Introduction to Stats, bulk RNAseq analysis, single cell RNAseq analysis, and ChIPseq analysis. We further have classes covering Methylation Arrays and GWAS (genome-wide association studies) in the development pipeline.
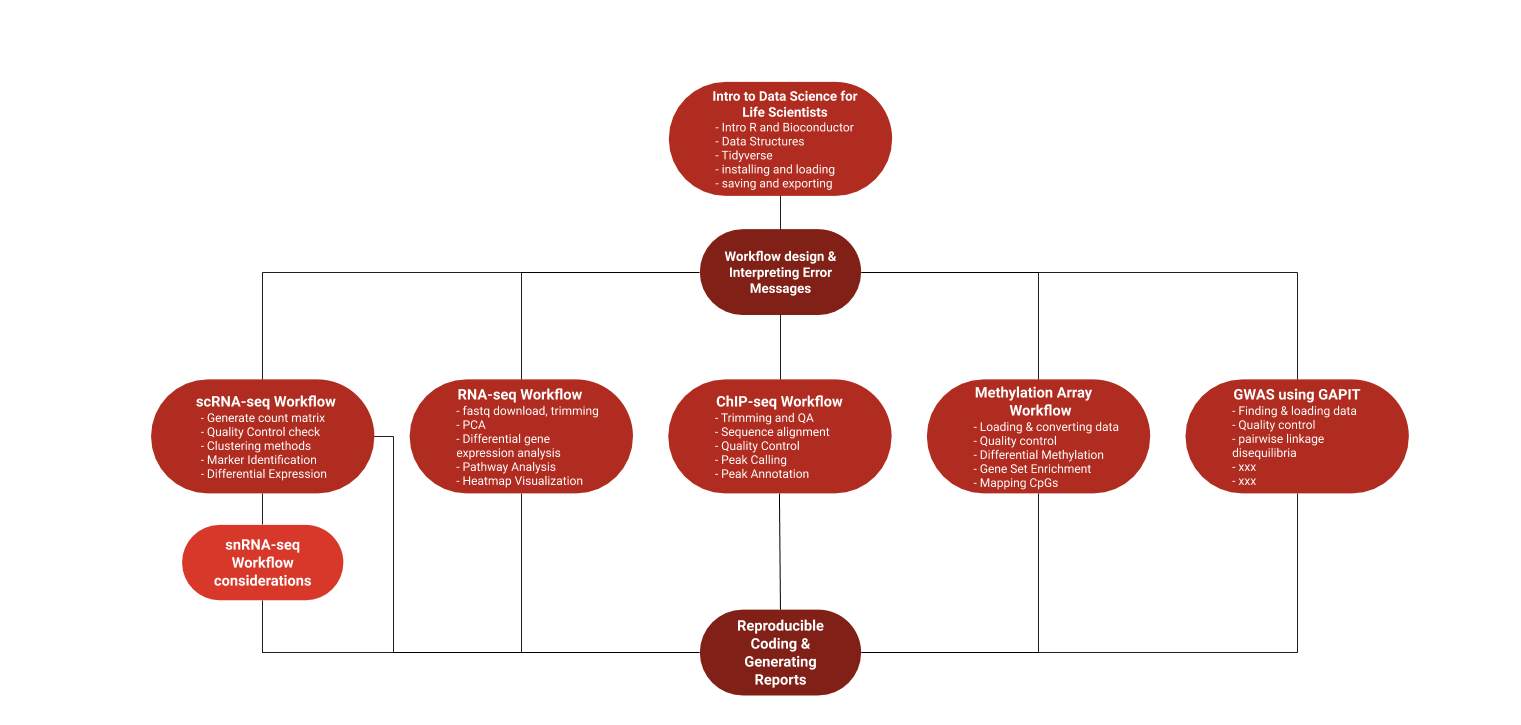
These courses provide interactive material that ranges from beginner's steps (package installation and environment setup) to low level, to high level analyses and visualizations. The courses are easily configurable and break down complex bioinformatics workflows into simple steps, and included standardized datasets to work with. Most importantly these interactive courses provide real time feedback on your work. Further, the material places strong emphasis on coding practices, open science, and reproducibility. Finally the whole framework as very easy to install and included datasets are small, so the courses work on low spec systems.
Have questions? Contact us at bioctoolsdevteam@gmail.com